Benchmark of SATLAS2 speed#
The simultaneous fit of sets of data is shown in this notebook. The data generation code can be replaced by code that reads in datafiles, so this script can serve as the basis for your own analysis. The fitting is compared to the simulatenous fit in the first version of satlas.
First, start with an import of all the relevant libraries:
import sys
import time
import matplotlib.gridspec as gridspec
import matplotlib.pyplot as plt
import numpy as np
sys.path.insert(0, '..\src')
import satlas2
import satlas as sat
Define a modified root function to handle uncertainties of 0 counts in a Poisson statistic
def modifiedSqrt(input):
output = np.sqrt(input)
output[input <= 0] = 1
return output
Define all the parameters and, in a for-loop, define the HFS and background models to generate the data from. The fitting already occurs inside the for loop, so the performance can be seen as a function of the number of datasets that are being analysed.
spin = 3.5
J = [0.5, 1.5]
A = [9600, 175]
B = [0, 315]
C = [0, 0]
FWHMG = 135
FWHML = 101
centroid = 480
bkg = 10
scale = 90
x = np.arange(-17500, -14500, 40)
x = np.hstack([x, np.arange(20000, 23000, 40)])
times = []
times_1 = []
rng = np.random.default_rng(0)
for j in range(1, 11):
f = satlas2.Fitter()
models = []
X = []
Y = []
for i in range(j):
hfs = satlas2.HFS(spin,
J,
A=A,
B=B,
C=C,
scale=scale,
df=centroid,
name='HFS1',
racah=True,
fwhmg=135,
fwhml=100)
bkgm = satlas2.Polynomial([bkg], name='bkg1')
y = satlas2.generateSpectrum([hfs, bkgm], x, rng.poisson)
hfs.params['centroid'].value = centroid - 100
X.append(x)
Y.append(y)
hfs1 = sat.HFSModel(spin,
J,
[A[0], A[1], B[0], B[1], C[0], C[1]],
centroid - 100, [FWHMG, FWHML],
scale=scale,
background_params=[bkg],
use_racah=True)
models.append(hfs1)
datasource = satlas2.Source(x,
y,
yerr=modifiedSqrt,
name='Scan{}'.format(i + 1))
datasource.addModel(hfs)
datasource.addModel(bkgm)
f.addSource(datasource)
share = ['Al', 'Au', 'Bl', 'centroid', 'FWHMG', 'FWHML']
m = sat.LinkedModel(models)
m.shared = share
f.shareModelParams(share)
print('Fitting {} datasets with chisquare (Pearson, satlas2)...'.format(j))
start = time.time()
f.fit()
stop = time.time()
dt = stop - start
print('{:.3} s, {:.0f} function evaluations'.format(dt, f.result.nfev))
times.append(dt)
print('Fitting {} datasets with chisquare (Pearson, satlas1)...'.format(j))
start = time.time()
sat.chisquare_spectroscopic_fit(m, X, Y)
stop = time.time()
dt = stop - start
times_1.append(dt)
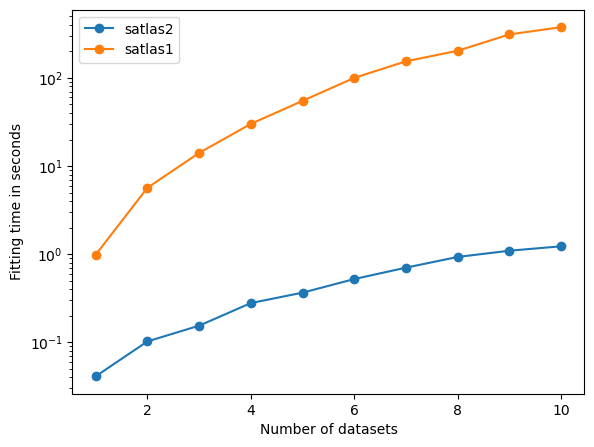
fig = plt.figure()
ax = fig.add_axes([0.1, 0.1, 0.8, 0.8])
ax.plot(range(1, len(times) + 1), times, '-o', label='satlas2')
ax.plot(range(1, len(times_1) + 1), times_1, '-o', label='satlas1')
ax.set_xlabel('Number of datasets')
ax.set_ylabel('Fitting time in seconds')
ax.set_yscale('log')
ax.legend(loc=0)
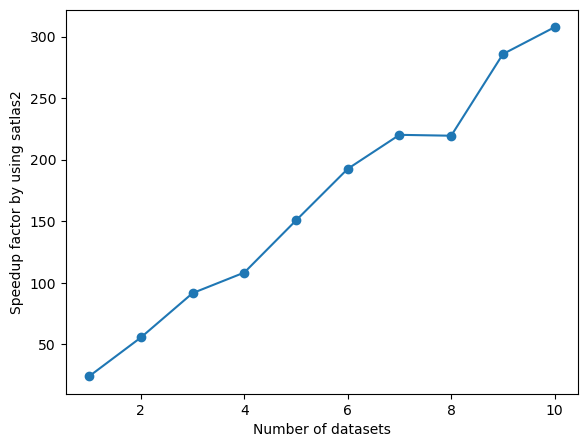
times, times_1 = np.array(times), np.array(times_1)
fig = plt.figure()
ax = fig.add_axes([0.1, 0.1, 0.8, 0.8])
ax.plot(range(1, len(times) + 1), times_1/times, '-o')
ax.set_xlabel('Number of datasets')
ax.set_ylabel('Speedup factor by using satlas2')
Fitting 1 datasets with chisquare (Pearson, satlas2)...
0.041 s, 73 function evaluations
Fitting 1 datasets with chisquare (Pearson, satlas1)...
Chisquare fitting done: 98it [00:00, 100.10it/s]
Fitting 2 datasets with chisquare (Pearson, satlas2)...
0.102 s, 110 function evaluations
Fitting 2 datasets with chisquare (Pearson, satlas1)...
Chisquare fitting done: 174it [00:05, 30.77it/s]
Fitting 3 datasets with chisquare (Pearson, satlas2)...
0.154 s, 122 function evaluations
Fitting 3 datasets with chisquare (Pearson, satlas1)...
Chisquare fitting done: 209it [00:14, 14.83it/s]
Fitting 4 datasets with chisquare (Pearson, satlas2)...
0.278 s, 163 function evaluations
Fitting 4 datasets with chisquare (Pearson, satlas1)...
Chisquare fitting in progress (516.8577280066263): 258it [00:29, 8.60it/s]
Fitting 5 datasets with chisquare (Pearson, satlas2)...
0.365 s, 169 function evaluations
Fitting 5 datasets with chisquare (Pearson, satlas1)...
Chisquare fitting in progress (791.4835074105964): 308it [00:54, 5.90it/s]
Fitting 6 datasets with chisquare (Pearson, satlas2)...
0.521 s, 217 function evaluations
Fitting 6 datasets with chisquare (Pearson, satlas1)...
Chisquare fitting in progress (921.0408291264894): 393it [01:39, 3.97it/s]
Fitting 7 datasets with chisquare (Pearson, satlas2)...
0.702 s, 244 function evaluations
Fitting 7 datasets with chisquare (Pearson, satlas1)...
Chisquare fitting in progress (1025.7328760442326): 448it [02:34, 2.88it/s]
Fitting 8 datasets with chisquare (Pearson, satlas2)...
0.929 s, 271 function evaluations
Fitting 8 datasets with chisquare (Pearson, satlas1)...
Chisquare fitting in progress (1116.8718639445108): 458it [03:23, 2.33it/s]
Fitting 9 datasets with chisquare (Pearson, satlas2)...
1.09 s, 298 function evaluations
Fitting 9 datasets with chisquare (Pearson, satlas1)...
Chisquare fitting in progress (1254.023933377538): 558it [05:11, 1.77it/s]
Fitting 10 datasets with chisquare (Pearson, satlas2)...
1.23 s, 290 function evaluations
Fitting 10 datasets with chisquare (Pearson, satlas1)...
Chisquare fitting in progress (1406.051401654012): 559it [06:16, 1.50it/s]
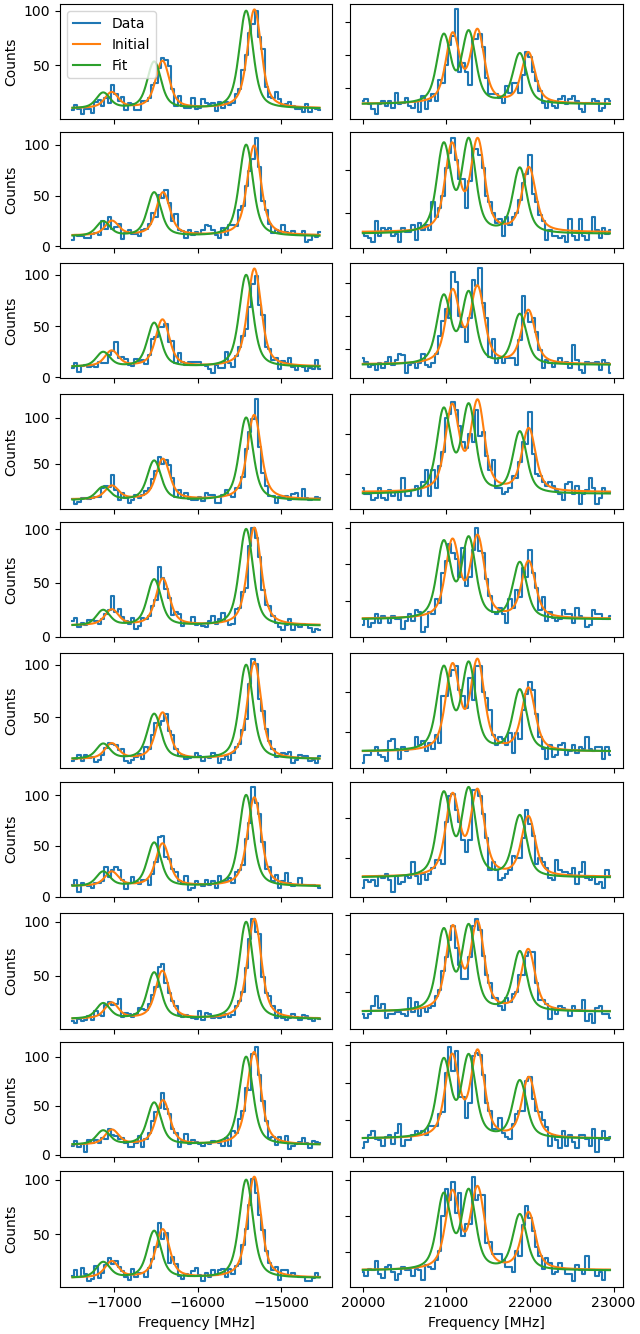
Plot the fit result, then revert the fit to show the initial starting condition of the spectrum.
fig = plt.figure(constrained_layout=True)
gs = gridspec.GridSpec(nrows=len(f.sources), ncols=2, figure=fig)
a1 = None
a2 = None
axes = []
for i, (name, datasource) in enumerate(f.sources):
if a1 is None:
ax1 = fig.add_subplot(gs[i, 0])
ax2 = fig.add_subplot(gs[i, 1])
a1 = ax1
a2 = ax2
else:
ax1 = fig.add_subplot(gs[i, 0], sharex=a1)
ax2 = fig.add_subplot(gs[i, 1], sharex=a2)
left = datasource.x < 0
right = datasource.x > 0
smooth_left = np.arange(datasource.x[left].min(), datasource.x[left].max(),
5.0)
smooth_right = np.arange(datasource.x[right].min(),
datasource.x[right].max(), 5.0)
ax1.plot(datasource.x[left],
datasource.y[left],
drawstyle='steps-mid',
label='Data')
ax1.plot(smooth_left, datasource.evaluate(smooth_left), label='Fit')
ax2.plot(datasource.x[right],
datasource.y[right],
drawstyle='steps-mid',
label='Data')
ax2.plot(smooth_right, datasource.evaluate(smooth_right), label='Fit')
ax1.set_xlabel('Frequency [MHz]')
ax2.set_xlabel('Frequency [MHz]')
ax1.set_ylabel('Counts')
ax2.set_ylabel('Counts')
ax1.label_outer()
ax2.label_outer()
axes.append([ax1, ax2])
f.revertFit()
for i, (name, datasource) in enumerate(f.sources):
smooth_left = np.arange(datasource.x[left].min(), datasource.x[left].max(),
5.0)
smooth_right = np.arange(datasource.x[right].min(),
datasource.x[right].max(), 5.0)
axes[i][0].plot(smooth_left, datasource.evaluate(smooth_left), label='Initial')
axes[i][1].plot(smooth_right,
datasource.evaluate(smooth_right),
label='Initial')
a1.legend(loc=0)
print(f.reportFit())
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 290
# data points = 1500
# variables = 35
chi-square = 1423.58804
reduced chi-square = 0.97173245
Akaike info crit = -8.42695240
Bayesian info crit = 177.535761
[[Variables]]
Scan1___HFS1___centroid: 481.497549 +/- 1.15593654 (0.24%) (init = 380)
Scan1___HFS1___Al: 9600.61046 +/- 0.92670540 (0.01%) (init = 9600)
Scan1___HFS1___Au: 174.571911 +/- 0.40166968 (0.23%) (init = 175)
Scan1___HFS1___Bl: 0 (fixed)
Scan1___HFS1___Bu: 316.727852 +/- 9.58185930 (3.03%) (init = 315)
Scan1___HFS1___Cl: 0 (fixed)
Scan1___HFS1___Cu: 0 (fixed)
Scan1___HFS1___FWHMG: 130.719040 +/- 8.12890265 (6.22%) (init = 135)
Scan1___HFS1___FWHML: 105.176292 +/- 7.66248618 (7.29%) (init = 100)
Scan1___HFS1___scale: 90.9386339 +/- 3.18982406 (3.51%) (init = 90)
Scan1___HFS1___Amp3to2: 0.4545455 (fixed)
Scan1___HFS1___Amp3to3: 0.4772727 (fixed)
Scan1___HFS1___Amp3to4: 0.3409091 (fixed)
Scan1___HFS1___Amp4to3: 0.1590909 (fixed)
Scan1___HFS1___Amp4to4: 0.4772727 (fixed)
Scan1___HFS1___Amp4to5: 1 (fixed)
Scan1___bkg1___p0: 10.2241495 +/- 0.38793282 (3.79%) (init = 10)
Scan2___HFS1___centroid: 481.497549 +/- 1.15593654 (0.24%) == 'Scan1___HFS1___centroid'
Scan2___HFS1___Al: 9600.61046 +/- 0.92670540 (0.01%) == 'Scan1___HFS1___Al'
Scan2___HFS1___Au: 174.571911 +/- 0.40166968 (0.23%) == 'Scan1___HFS1___Au'
Scan2___HFS1___Bl: 0.00000000 +/- 0.00000000 == 'Scan1___HFS1___Bl'
Scan2___HFS1___Bu: 301.516120 +/- 9.76476582 (3.24%) (init = 315)
Scan2___HFS1___Cl: 0 (fixed)
Scan2___HFS1___Cu: 0 (fixed)
Scan2___HFS1___FWHMG: 130.719040 +/- 8.12890268 (6.22%) == 'Scan1___HFS1___FWHMG'
Scan2___HFS1___FWHML: 105.176292 +/- 7.66248618 (7.29%) == 'Scan1___HFS1___FWHML'
Scan2___HFS1___scale: 88.4215797 +/- 3.18866686 (3.61%) (init = 90)
Scan2___HFS1___Amp3to2: 0.4545455 (fixed)
Scan2___HFS1___Amp3to3: 0.4772727 (fixed)
Scan2___HFS1___Amp3to4: 0.3409091 (fixed)
Scan2___HFS1___Amp4to3: 0.1590909 (fixed)
Scan2___HFS1___Amp4to4: 0.4772727 (fixed)
Scan2___HFS1___Amp4to5: 1 (fixed)
Scan2___bkg1___p0: 10.7465561 +/- 0.39604567 (3.69%) (init = 10)
Scan3___HFS1___centroid: 481.497549 +/- 1.15593654 (0.24%) == 'Scan1___HFS1___centroid'
Scan3___HFS1___Al: 9600.61046 +/- 0.92670540 (0.01%) == 'Scan1___HFS1___Al'
Scan3___HFS1___Au: 174.571911 +/- 0.40166968 (0.23%) == 'Scan1___HFS1___Au'
Scan3___HFS1___Bl: 0.00000000 +/- 0.00000000 == 'Scan1___HFS1___Bl'
Scan3___HFS1___Bu: 316.467273 +/- 9.15709217 (2.89%) (init = 315)
Scan3___HFS1___Cl: 0 (fixed)
Scan3___HFS1___Cu: 0 (fixed)
Scan3___HFS1___FWHMG: 130.719040 +/- 8.12890268 (6.22%) == 'Scan1___HFS1___FWHMG'
Scan3___HFS1___FWHML: 105.176292 +/- 7.66248618 (7.29%) == 'Scan1___HFS1___FWHML'
Scan3___HFS1___scale: 95.8064722 +/- 3.27951355 (3.42%) (init = 90)
Scan3___HFS1___Amp3to2: 0.4545455 (fixed)
Scan3___HFS1___Amp3to3: 0.4772727 (fixed)
Scan3___HFS1___Amp3to4: 0.3409091 (fixed)
Scan3___HFS1___Amp4to3: 0.1590909 (fixed)
Scan3___HFS1___Amp4to4: 0.4772727 (fixed)
Scan3___HFS1___Amp4to5: 1 (fixed)
Scan3___bkg1___p0: 10.3773605 +/- 0.39449044 (3.80%) (init = 10)
Scan4___HFS1___centroid: 481.497549 +/- 1.15593654 (0.24%) == 'Scan1___HFS1___centroid'
Scan4___HFS1___Al: 9600.61046 +/- 0.92670540 (0.01%) == 'Scan1___HFS1___Al'
Scan4___HFS1___Au: 174.571911 +/- 0.40166968 (0.23%) == 'Scan1___HFS1___Au'
Scan4___HFS1___Bl: 0.00000000 +/- 0.00000000 == 'Scan1___HFS1___Bl'
Scan4___HFS1___Bu: 306.363833 +/- 9.44073795 (3.08%) (init = 315)
Scan4___HFS1___Cl: 0 (fixed)
Scan4___HFS1___Cu: 0 (fixed)
Scan4___HFS1___FWHMG: 130.719040 +/- 8.12890268 (6.22%) == 'Scan1___HFS1___FWHMG'
Scan4___HFS1___FWHML: 105.176292 +/- 7.66248618 (7.29%) == 'Scan1___HFS1___FWHML'
Scan4___HFS1___scale: 91.9771725 +/- 3.22329550 (3.50%) (init = 90)
Scan4___HFS1___Amp3to2: 0.4545455 (fixed)
Scan4___HFS1___Amp3to3: 0.4772727 (fixed)
Scan4___HFS1___Amp3to4: 0.3409091 (fixed)
Scan4___HFS1___Amp4to3: 0.1590909 (fixed)
Scan4___HFS1___Amp4to4: 0.4772727 (fixed)
Scan4___HFS1___Amp4to5: 1 (fixed)
Scan4___bkg1___p0: 10.8933956 +/- 0.39725280 (3.65%) (init = 10)
Scan5___HFS1___centroid: 481.497549 +/- 1.15593654 (0.24%) == 'Scan1___HFS1___centroid'
Scan5___HFS1___Al: 9600.61046 +/- 0.92670540 (0.01%) == 'Scan1___HFS1___Al'
Scan5___HFS1___Au: 174.571911 +/- 0.40166968 (0.23%) == 'Scan1___HFS1___Au'
Scan5___HFS1___Bl: 0.00000000 +/- 0.00000000 == 'Scan1___HFS1___Bl'
Scan5___HFS1___Bu: 311.300307 +/- 9.57352553 (3.08%) (init = 315)
Scan5___HFS1___Cl: 0 (fixed)
Scan5___HFS1___Cu: 0 (fixed)
Scan5___HFS1___FWHMG: 130.719040 +/- 8.12890268 (6.22%) == 'Scan1___HFS1___FWHMG'
Scan5___HFS1___FWHML: 105.176292 +/- 7.66248618 (7.29%) == 'Scan1___HFS1___FWHML'
Scan5___HFS1___scale: 90.7998344 +/- 3.20100095 (3.53%) (init = 90)
Scan5___HFS1___Amp3to2: 0.4545455 (fixed)
Scan5___HFS1___Amp3to3: 0.4772727 (fixed)
Scan5___HFS1___Amp3to4: 0.3409091 (fixed)
Scan5___HFS1___Amp4to3: 0.1590909 (fixed)
Scan5___HFS1___Amp4to4: 0.4772727 (fixed)
Scan5___HFS1___Amp4to5: 1 (fixed)
Scan5___bkg1___p0: 10.3707148 +/- 0.39092416 (3.77%) (init = 10)
Scan6___HFS1___centroid: 481.497549 +/- 1.15593654 (0.24%) == 'Scan1___HFS1___centroid'
Scan6___HFS1___Al: 9600.61046 +/- 0.92670540 (0.01%) == 'Scan1___HFS1___Al'
Scan6___HFS1___Au: 174.571911 +/- 0.40166968 (0.23%) == 'Scan1___HFS1___Au'
Scan6___HFS1___Bl: 0.00000000 +/- 0.00000000 == 'Scan1___HFS1___Bl'
Scan6___HFS1___Bu: 313.188923 +/- 9.22636900 (2.95%) (init = 315)
Scan6___HFS1___Cl: 0 (fixed)
Scan6___HFS1___Cu: 0 (fixed)
Scan6___HFS1___FWHMG: 130.719040 +/- 8.12890268 (6.22%) == 'Scan1___HFS1___FWHMG'
Scan6___HFS1___FWHML: 105.176292 +/- 7.66248618 (7.29%) == 'Scan1___HFS1___FWHML'
Scan6___HFS1___scale: 92.7961475 +/- 3.20546516 (3.45%) (init = 90)
Scan6___HFS1___Amp3to2: 0.4545455 (fixed)
Scan6___HFS1___Amp3to3: 0.4772727 (fixed)
Scan6___HFS1___Amp3to4: 0.3409091 (fixed)
Scan6___HFS1___Amp4to3: 0.1590909 (fixed)
Scan6___HFS1___Amp4to4: 0.4772727 (fixed)
Scan6___HFS1___Amp4to5: 1 (fixed)
Scan6___bkg1___p0: 9.85910281 +/- 0.38300602 (3.88%) (init = 10)
Scan7___HFS1___centroid: 481.497549 +/- 1.15593654 (0.24%) == 'Scan1___HFS1___centroid'
Scan7___HFS1___Al: 9600.61046 +/- 0.92670540 (0.01%) == 'Scan1___HFS1___Al'
Scan7___HFS1___Au: 174.571911 +/- 0.40166968 (0.23%) == 'Scan1___HFS1___Au'
Scan7___HFS1___Bl: 0.00000000 +/- 0.00000000 == 'Scan1___HFS1___Bl'
Scan7___HFS1___Bu: 315.004090 +/- 10.1665755 (3.23%) (init = 315)
Scan7___HFS1___Cl: 0 (fixed)
Scan7___HFS1___Cu: 0 (fixed)
Scan7___HFS1___FWHMG: 130.719040 +/- 8.12890268 (6.22%) == 'Scan1___HFS1___FWHMG'
Scan7___HFS1___FWHML: 105.176292 +/- 7.66248618 (7.29%) == 'Scan1___HFS1___FWHML'
Scan7___HFS1___scale: 87.2691437 +/- 3.12308461 (3.58%) (init = 90)
Scan7___HFS1___Amp3to2: 0.4545455 (fixed)
Scan7___HFS1___Amp3to3: 0.4772727 (fixed)
Scan7___HFS1___Amp3to4: 0.3409091 (fixed)
Scan7___HFS1___Amp4to3: 0.1590909 (fixed)
Scan7___HFS1___Amp4to4: 0.4772727 (fixed)
Scan7___HFS1___Amp4to5: 1 (fixed)
Scan7___bkg1___p0: 10.3964797 +/- 0.38718000 (3.72%) (init = 10)
Scan8___HFS1___centroid: 481.497549 +/- 1.15593654 (0.24%) == 'Scan1___HFS1___centroid'
Scan8___HFS1___Al: 9600.61046 +/- 0.92670540 (0.01%) == 'Scan1___HFS1___Al'
Scan8___HFS1___Au: 174.571911 +/- 0.40166968 (0.23%) == 'Scan1___HFS1___Au'
Scan8___HFS1___Bl: 0.00000000 +/- 0.00000000 == 'Scan1___HFS1___Bl'
Scan8___HFS1___Bu: 319.167680 +/- 9.49188859 (2.97%) (init = 315)
Scan8___HFS1___Cl: 0 (fixed)
Scan8___HFS1___Cu: 0 (fixed)
Scan8___HFS1___FWHMG: 130.719040 +/- 8.12890268 (6.22%) == 'Scan1___HFS1___FWHMG'
Scan8___HFS1___FWHML: 105.176292 +/- 7.66248618 (7.29%) == 'Scan1___HFS1___FWHML'
Scan8___HFS1___scale: 92.6245328 +/- 3.20556643 (3.46%) (init = 90)
Scan8___HFS1___Amp3to2: 0.4545455 (fixed)
Scan8___HFS1___Amp3to3: 0.4772727 (fixed)
Scan8___HFS1___Amp3to4: 0.3409091 (fixed)
Scan8___HFS1___Amp4to3: 0.1590909 (fixed)
Scan8___HFS1___Amp4to4: 0.4772727 (fixed)
Scan8___HFS1___Amp4to5: 1 (fixed)
Scan8___bkg1___p0: 10.1201224 +/- 0.38736226 (3.83%) (init = 10)
Scan9___HFS1___centroid: 481.497549 +/- 1.15593654 (0.24%) == 'Scan1___HFS1___centroid'
Scan9___HFS1___Al: 9600.61046 +/- 0.92670540 (0.01%) == 'Scan1___HFS1___Al'
Scan9___HFS1___Au: 174.571911 +/- 0.40166968 (0.23%) == 'Scan1___HFS1___Au'
Scan9___HFS1___Bl: 0.00000000 +/- 0.00000000 == 'Scan1___HFS1___Bl'
Scan9___HFS1___Bu: 303.519268 +/- 9.25628071 (3.05%) (init = 315)
Scan9___HFS1___Cl: 0 (fixed)
Scan9___HFS1___Cu: 0 (fixed)
Scan9___HFS1___FWHMG: 130.719040 +/- 8.12890268 (6.22%) == 'Scan1___HFS1___FWHMG'
Scan9___HFS1___FWHML: 105.176292 +/- 7.66248618 (7.29%) == 'Scan1___HFS1___FWHML'
Scan9___HFS1___scale: 94.8212808 +/- 3.23148508 (3.41%) (init = 90)
Scan9___HFS1___Amp3to2: 0.4545455 (fixed)
Scan9___HFS1___Amp3to3: 0.4772727 (fixed)
Scan9___HFS1___Amp3to4: 0.3409091 (fixed)
Scan9___HFS1___Amp4to3: 0.1590909 (fixed)
Scan9___HFS1___Amp4to4: 0.4772727 (fixed)
Scan9___HFS1___Amp4to5: 1 (fixed)
Scan9___bkg1___p0: 9.99299420 +/- 0.38760703 (3.88%) (init = 10)
Scan10___HFS1___centroid: 481.497549 +/- 1.15593654 (0.24%) == 'Scan1___HFS1___centroid'
Scan10___HFS1___Al: 9600.61046 +/- 0.92670540 (0.01%) == 'Scan1___HFS1___Al'
Scan10___HFS1___Au: 174.571911 +/- 0.40166968 (0.23%) == 'Scan1___HFS1___Au'
Scan10___HFS1___Bl: 0.00000000 +/- 0.00000000 == 'Scan1___HFS1___Bl'
Scan10___HFS1___Bu: 311.540881 +/- 9.35397017 (3.00%) (init = 315)
Scan10___HFS1___Cl: 0 (fixed)
Scan10___HFS1___Cu: 0 (fixed)
Scan10___HFS1___FWHMG: 130.719040 +/- 8.12890268 (6.22%) == 'Scan1___HFS1___FWHMG'
Scan10___HFS1___FWHML: 105.176292 +/- 7.66248618 (7.29%) == 'Scan1___HFS1___FWHML'
Scan10___HFS1___scale: 92.4534513 +/- 3.22161005 (3.48%) (init = 90)
Scan10___HFS1___Amp3to2: 0.4545455 (fixed)
Scan10___HFS1___Amp3to3: 0.4772727 (fixed)
Scan10___HFS1___Amp3to4: 0.3409091 (fixed)
Scan10___HFS1___Amp4to3: 0.1590909 (fixed)
Scan10___HFS1___Amp4to4: 0.4772727 (fixed)
Scan10___HFS1___Amp4to5: 1 (fixed)
Scan10___bkg1___p0: 10.2348292 +/- 0.39038996 (3.81%) (init = 10)